Research Interest
Our research program involves a multi-disciplinary approach and many collaborations. Our interests range from understanding molecular mechanisms to translating these findings to address leading biomedical problems.
The discovery of new potential therapeutic targets has thus far outpaced the translation of these findings into innovative therapy. Currently, the portfolio of FDA-approved drugs targets only a tiny fraction of the human proteome for therapies for all human diseases. Signal transduction pathways that are regulated by protein post-translational modifications, such as phosphorylation and acetylation, are proven therapeutic approaches of human diseases. Although significant information has been obtained in targeting protein phosphorylation, acetylation and methylation, much less knowledge exists in modulating the largest family of enzymes that catalyze a major type of post-translational modifications – ubiquitin-like (Ubl) modifications. Our lab has made a major contribution to our understanding of enzymatic mechanism of Ubl modifications. Ubl modifications involve protein substrates and multiple steps. We developed quantitative approaches to examine the enzyme kinetics of every step of the multi-step and multi-enzyme conjugation process of Ubl modification, which is more complex than any other post-translational modification, such as phosphorylation. The expertise and reagents developed have been critical in the discovery and development of inhibitors to target these enzymes for developing potential therapeutics. Our representative recent work includes identification of a small molecular inhibitor of small ubiquitin-like modifications through a combination of high throughput screening followed by enzymological characterization, and elucidation of the allosteric inhibition mechanism (Cell Chemical Biology, 26:278-288.e6, 2019; Nature Communications, 9:5145, 2018).
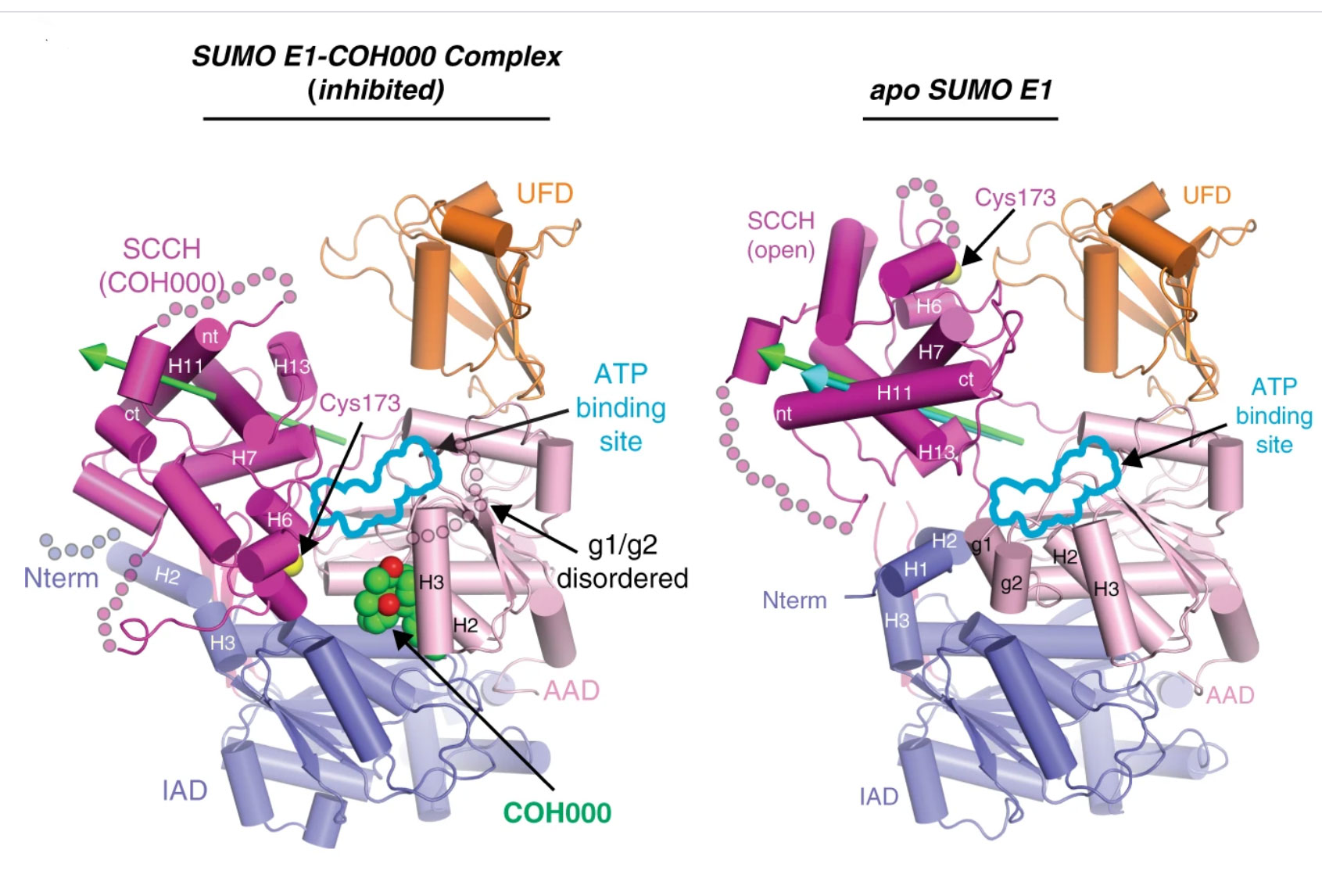
Figure Legend: Crystal structures showing how the first-in-class allosteric inhibitor COH000 completely inhibits SUMO activating enzyme (SAE) activity. The green and cyan arrows highlight the rotation axes during transition of the SCCH domain from open to COH000-inhibited state and conformational change required for its catalytic cycle, respectively.

Figure Legend: Zoom in view of the binding pocket for COH000 in SAE.
A long-term interest of our group is to elucidate how SUMOylation regulates cellular functions. We identified the first SUMO-interacting motif (SIM) that mediates all SUMO-dependent protein-protein interactions (Proceedings of the National Academy of Sciences, 101:14373-14378, 2004). We also have shown that SUMOylation is a target for inhibiting cancer stem cells (Nature Communications 7: 12326, 2016). Cancer stem cells (CSCs) have key roles in treatment resistance, tumor metastasis and relapse. Using colorectal cancer (CC) cell lines and patient-derived xenograft (PDX) tissues, we found that CC CSCs, which resist chemoradiation, have higher SAE and global SUMOylation levels than non-CSCs. Knockdown of SAE or SUMO conjugating enzyme (Ubc9) inhibits CC CSC maintenance and self-renewal, while overexpression of SAE or Ubc9 increases CC cell stemness. We found that SUMOylation regulates CSCs through Oct-1, a transcription factor for aldehyde dehydrogenases (ALDHs). ALDH activity is not only a marker for CSCs but also important in CSC biology. SUMO does not modify Oct-1 directly, but regulates the expression of TRIM21 that enhances Oct-1 ubiquitination and, consequently, reducing Oct-1 stability. In summary, our findings suggest that SUMOylation could be a target to inhibit CSCs and ultimately to reduce treatment resistance, tumor metastasis and relapse.
More recently, we elucidated how SUMOylation regulates miR-34b/c-targeted gene expression program (Nucleic Acids Research. 46: 7108–7123, 2018).

Figure legend: (a) Representative colony formation assay of ALDH+ cells transduced with control non-silencing shRNA (shCtrl) or shSAE2 lentivirus and grown in Matrigel (scale bar, 20 μm). (b,c) Quantification of colony number (b) and colony size (c) from four different wells and six fields in each well of the experiments described in a. (d,e) Frequency of CSC in HT29 ALDH+ cells transduced with shCtrl or shSAE2 lentivirus measured by LDA in vivo as shown by the detailed data (d) and as a plot of fold changes in the CSC frequency (e). (f) Tumor growth curve for HT29 ALDH+ shCtrl or shSAE2 cells injected subcutaneously with 500 cells per mouse of six mice in each group. *P<0.05, **P<0.01 and ***P<0.001.
Nature Communications 7: 12326, 2016

Figure legend: miR-34b/c expression is SUMOylation-dependent. (A, B) Pri-miR-34b/c (A) and mature miR-34b/c (B) were measured using qRT-PCR after either siRNA knockdown or ectopic expression of Ubc9 or SAE2 in HCT116 and LCL cells. Expression of pri-miR-34b/c was first normalized against GAPDH and then to the control (siCtrl). Expression of mature miR-34b/c was first normalized against RNU6B and then to the control. (C) Chromatin immunoprecipitation assay (ChIP) of transcription factors known or predicted to bind the miR-34b/c promoter without (DOX–) or with (DOX+) SAE2 knockdown. The results were normalized to total DNA input. IgG was used as a control. (D) Nuclear localization of FOXO3a increased after SAE2 knockdown (DOX+). Cell fractionation was carried out to separate cytoplasm (C) and nuclei (N) fractions and then Western blot was used to detect FOXO3a, phosphorylated FOXO3a (p-FOXO3a, Ser253), GAPDH, and histone H3. (E) FOXO3a localization was also visualized using IF. DAPI staining shows nuclei. The white bar represents 10 μm. All results shown as mean ± STDEV from three independent experiments. Statistical significance of the data was analyzed using one-way ANOVA. **P < 0.01; ***P < 0.001.

Figure legend: Mutation of SUMO acceptor lysines results in hyperactive NLRP3. a–c Immunoblot analysis of inflammasome components (caspase-1, ASC and NLRP3) in cell lysates of HEK239T cells, stably expressing ASC and caspase-1, transfected with the indicated proteins. Caspase-1 maturation (p20 band) was evaluated by western blot. d NMR analysis of the interaction between UBC9 and WT and K689R mutant NLRP3 peptides. Overlays of the expanded regions of 1H-15N HSQC spectra of free UBC9 (red) and UBC9 in complex with the peptide at 8:1 molar ratio (green). Only peaks with significant chemical shift perturbation are labelled with their assignments. e In vitro sumoylation assays of WT and mutant peptides using SUMO-1 or GST-fusion SUMO-3. f HEK293T cells were transfected with plasmids expressing the indicated proteins and FLAG-NLRP3 immunoprecipitates were evaluated for sumoylation and ubiquitylation. The modifications were quantified using Image Lab Software (Bio-Rad) and normalised to total immunoprecipitated NLRP3. The data of three independent experiments are presented as log2-transformed fold changes of the mutants compared to WT NLRP3. Line represents the mean. *P < 0.05; two-tailed Student’s t-test comparing each individual mutant to WT NLRP3.
Our recent focus has become anti-tumor immune response that resulted in fruitful publications. In collaboration with Dr. Pascal Meier (Institute of Cancer Research, UK), we investigated the role of SUMOylation in NLRP3 inflammasome (Nature Communications, 9: 3001, 2018), which responds to infection and tissue damage, and rapidly escalates the intensity of inflammation. We show that NLRP3 inflammasome activation is suppressed by SUMOylation. NLRP3 is SUMOylated by the SUMO E3-ligase MAPL. Defective NLRP3 SUMOylation results in enhanced caspase-1 activation and IL-1β release.
In collaboration with Dr. Zuoming Sun’s laboratory, we showed that the function of transcription factor RORγt is regulated by its SUMOylation (Nature Communications, 9:4870, 2018). Loss of SUMO3, but not SUMO1, dampens TH17 differentiation and delays the progression of thymic CD8+ immature single-positive cells (ISPs) through reduced expression of IL17A. Mechanistically, SUMOylation of RORγt recruits histone acetyltransferase KAT2A, which stabilizes the binding of SRC1 to enhance RORγt transcription factor activity. This study demonstrates that SUMOylation is a critical mechanism for regulating RORγt function.
Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, Paladino D, Zhao J, Chen Y, Gunning PT, Turkson J.
Proc Natl Acad Sci U S A. 109(24):9623-8, 2012.
Targeted inhibition of TET1 transcription as a potent therapeutic strategy for acute myeloid leukemia. Jiang X, Hu C, Ferchen K, Nie J, Cui X, Chen C-H, Zuo Z, Seibel W, Skibbe JR, Cheng L, Tang Y, Wunderlich M, Reinhold WC, Arnovitz S, Ulrich B, Lu J, Weng H, Su R, Huang H, Dong L, Wang Y, Li C, Qin X, Mulloy J, Zheng Y, Diao J, Jin J, Li C, Liu PL, He C, Chen Y, Chen J.
Nature Communications., 8(1):2099, 2017.
